Readme from the project
Diffusion profile realignment (dpr)
An example and assorted implementation from the manuscript Reducing variability in along-tract analysis with diffusion profile realignment. Have a look at the example on how to use the package and run it on a small example dataset.
To install the package, just run
pip install dpr
The documentation is available at https://dpr.readthedocs.io.
The matlab version
There is also a shiny new version written in matlab, with an assorted example, available in the matlab folder. Feel free to check out and edit the functions as needed for your own usage.
A quick example from the command line
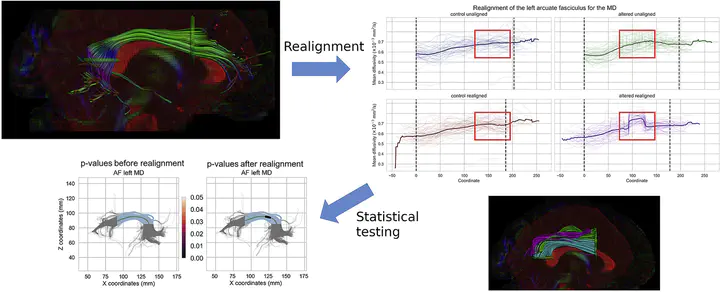
There is also a command line version for easy usage, here in an example for the AFD metric on the left arcuate fasciculus. The text file is already ordered in increasing order for each subject, which have the same distance between every point and are already zero padded accordingly.
We also supply the –exploredti option to remove the header column, –do_graph to save a png file with the results.
We finally resample everything to 75 points with –points 75.
The -f option overwrites the output files and the -v option prints useful informative messages throughout (and are optional).
dpr datasets/af_left_AFD.txt datasets/af_left_AFD_realigned.txt --exploredti --do_graph -f -v --points 75
The output datasets/af_left_AFD_realigned.txt is a text file where each line is a subject and each column is a different point of the along tract analysis.
We also get a png file datasets/af_left_AFD_realigned.png with the before/after realignment process.

Note how the zero padding present in the original data is decreasing the metrics as less and less subjects are present. The realigned metric is instead using padding with Nans, remember to consider/keep track of it in subsequent analysis as needed.
Visualizing the results
We can also draw the p-values (computed separately) over the bundle using the script dpr_make_fancy_graph.
This requires the original coordinates, the truncated version between rois and the coordinates to the representative streamline.
dpr_make_fancy_graph datasets/af_left_pval_unaligned.txt datasets/af_left_coordinates.txt datasets/af_left_truncated_coordinates.txt datasets/af_left_average_coordinates.txt 0,2 pvals_unaligned.png --title 'p-values before realignment' -f
dpr_make_fancy_graph datasets/af_left_pval_realigned.txt datasets/af_left_coordinates.txt datasets/af_left_truncated_coordinates.txt datasets/af_left_average_coordinates.txt 0,2 pvals_realigned.png -f
And this is the results


Using docker (instead of installing everything)
Well, you will still need to install docker first, see https://runnable.com/docker/getting-started/. After that, you can now pull a version from Dockerhub.
docker pull samuelstjean/dpr:v0.2
The two diffusion profile realignment scripts can now be run using
docker run samuelstjean/dpr:v0.2 # the default command is to run dpr --help
docker run samuelstjean/dpr:v0.2 dpr_make_fancy_graph --help
That’s it for the basic version, don’t hesitate to adapt everything to your needs.
As a more advanced example, to mount your own data folder inside the container (here we re-use the previous example), use
docker run -it -v /home/samuel/git/dpr/datasets:/mnt samuelstjean/dpr:v0.2 dpr /mnt/af_left_AFD.txt /mnt/af_left_AFD_realigned.txt --exploredti --do_graph -f -v --points 75
docker run -it -v /home/samuel/git/dpr/datasets:/mnt samuelstjean/dpr:v0.2 dpr_make_fancy_graph /mnt/af_left_pval_unaligned.txt /mnt/af_left_coordinates.txt /mnt/af_left_truncated_coordinates.txt /mnt/af_left_average_coordinates.txt 0,2 /mnt/pvals_unaligned.png --title 'p-values before realignment' -f
The result will be located in the folder you specified, here we used /home/samuel/git/dpr/datasets.
Datasets and reference
The main reference is
Samuel St-Jean, Maxime Chamberland, Max A. Viergever, Alexander Leemans,
Reducing variability in along-tract analysis with diffusion profile realignment,
NeuroImage, 2019. ISSN 1053-811
The data is also available at https://zenodo.org/record/2483169.
The open access manuscript is also available at https://www.sciencedirect.com/science/article/pii/S1053811919305014.
To refer to a specific version of the code, everything is also archived over at https://zenodo.org/record/3248894.
